Cucumbers, a summer favourite in salads and sandwiches, are not only an important commercial crop but also valuable model plants advancing genomic research.
A collaborative study between the John Innes Centre and the Chinese Academy of Agricultural Sciences (CAAS) combined genomic analysis and experiments to explore molecular differences between wild cucumbers and their domesticated counterparts.
Focusing on the genetics behind fruit elongation — since cultivated cucumbers are longer than their shorter, bitter wild relatives — the team uncovered insights that could enable more precise breeding of larger, higher-yielding crops with greater diversity.
Traditional plant breeding often targets mutations in protein-coding DNA sequences, which control traits like fruit size, flavour, and seed shape. However, these genes represent only a small fraction of the genome. Increasingly, researchers are turning to non-coding DNA regions once considered “silent.”
Synonymous mutations — formerly called silent mutations — occur in non-coding regions and are gaining attention for their role in cellular function. While previous studies hinted at their importance, evidence of their influence on traits in complex organisms has been limited, according to a press release.
Published in Cell, this study reveals how a single synonymous mutation influences cucumber fruit length by altering RNA structure and function — a molecule essential in gene expression.
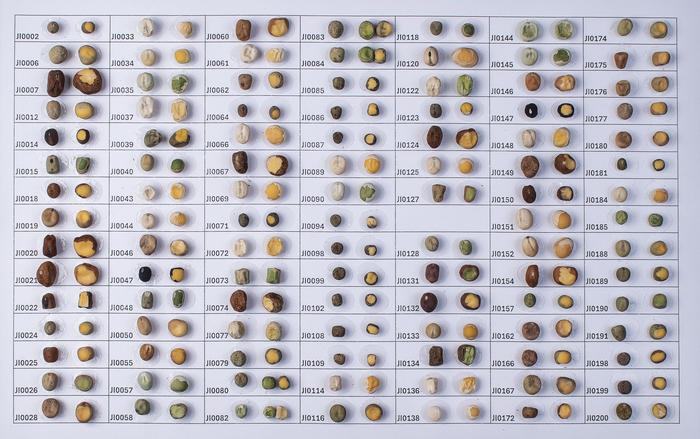
Using a genomic variation map of cucumber populations, the researchers identified fruit length as a key domestication trait. Molecular and genetic analyses showed that this single mutation was a major driver of fruit elongation during domestication, resulting in fruits up to 70% longer.
Importantly, this mutation doesn’t affect protein production directly, as is common with most agriculturally important traits. Instead, it reshapes RNA, which in turn suppresses the protein responsible for the shorter fruit trait found in wild cucumbers.
“A tiny ‘silent’ change in a cucumber gene — once thought to be innocuous — is the key player in making modern cucumbers longer,” said Dr Yueying Zhang, postdoctoral researcher at the John Innes Centre and first author of the study.
“Remarkably this silent mutation, long thought to be biologically neutral, rewired RNA regulation and contributed directly to the development of a domesticated trait.”
The findings offer valuable insights for crop breeding programs, presenting new opportunities to engineer important traits in the future. This research is particularly significant for traits like fruit size, which directly impact crop yield and provide commercial benefits for growers.
It also opens the door for further studies focused on synonymous sites, leveraging precision breeding tools such as gene editing to enhance traits across a wide range of crops.
This pioneering research was a collaborative effort between Professor Yiliang Ding’s team at the John Innes Centre, UK, and groups led by Professor Xueyong Yang at the Institute of Vegetables and Flowers, along with Academician Sanwen Huang, FRS, President of the Chinese Academy of Agricultural Sciences (CAAS). The project was initiated and led by CAAS, with Professor Ding’s team providing key expertise on RNA structure and translation regulation.