Promising techniques to accelerate innovation in plant breeding.
It has been recited many times before, our planet is faced with some of the most fearsome challenges it has ever seen. We need to produce more food and more energy for an ever-growing population, and we need to do that on less land, with less water, less resources, and in a more sustainable manner. And all of that in a changing climate.
Over the past century, plant breeding has been a major contributor, improving plant varieties to cope with population growth. However, due to urbanization, agriculture has been pushed to ever more marginal lands, and yield increases have been plateauing in several crops. So plant breeders will need to step up their efforts. Continuing on the way they have done so far won’t suffice in the coming decades. We need another revolution. And this revolution may very well come in the form of new breeding techniques (NBTs).
This group of techniques has been developed over the past 10 to 15 years, both in the public as well as in the private sector. The beauty of these techniques is they are capable of delivering a desired genetic trait(s) in a much more precise way than other techniques could, so far. Whereas in current plant breeding there are sometimes limitations in delivering the right characteristics to the target varieties, these techniques offer new possibilities to EU plant breeders.
It is a known fact that conventional plant breeding takes time. Surveys among plant breeding companies show it can take, on average, from seven to 12 (sometimes up to 20) years to generate a new plant variety with the desired characteristics, depending on the crop. Use of NBTs significantly shortens this period.
For example, in certain species it can be very time consuming to introduce a new resistance gene from the same, or related, species, due to the crop’s complex genetics. The result is, not all crosses produce fertile offspring. In addition, the growth habit of the crop itself can prevent a quick introduction of the trait. For example, trees take several years until the first flowers and fruits develop, and it can take decades to create a new variety. In this article, we’ll provide a technical overview of the different methods developed so far. A future issue of European Seed will address the regulatory environment surrounding NBTs.
Sequence-Specific Nuclease Technology
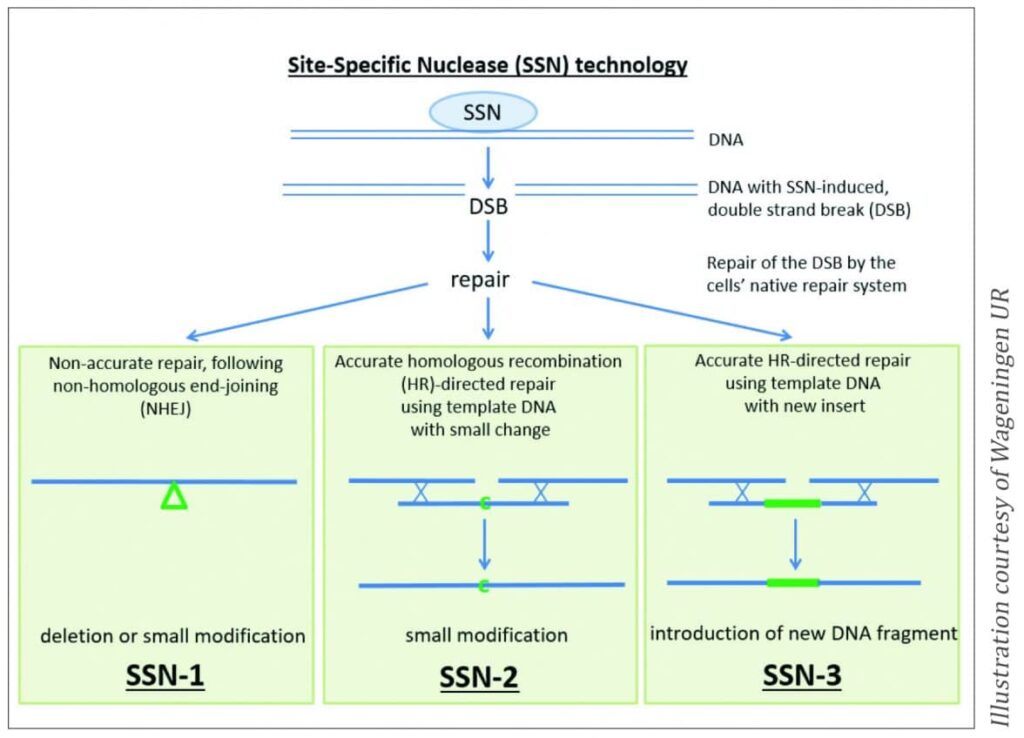
Sequence-specific nuclease (SSN) technology is often referred to as site-directed nuclease. It uses natural enzymes that generate a double-strand break in the DNA. These enzymes are linked to man-made structures designed to bind to a specific target DNA sequence. The complex causes a break at an exact pre-defined location in the DNA. The plant’s own repair mechanism repairs the break, but often inaccurately. There are three application types of SSN, SSN-1, SSN-2 and SSN-3.
With the application type SSN-1, no donor-DNA is used to guide the repair. Non-homologous end-joining (NHEJ) takes place, resulting, in most instances, in small deletions in the DNA, however, sometimes small additions can take place. These small alterations lead to loss of gene function (a gene knock-out).
The SSN-2 technique uses a donor DNA, which is a copy of the target DNA region with a small modification. During repair, the plant will use this template for the repair, and the small modification will be introduced into the plant’s genome (targeted mutation).
The repair template of the SSN-3 application type contains a complete new gene. Using SSN-3, intragenes, cisgenes (see below) or transgenes can be introduced (gene addition).
In any of the three ways described above, with SSN, a gene of interest can be mutated, replaced or knocked out (Figure 1). CRISPR-Cas9, zinc-finger nucleases (ZFNs), TALENs and meganucleases are all different variants of SSN.
For decades, plant breeders have been using classical mutagenesis methods, such as chemicals or ionizing radiation. In a way, similar results can be obtained with SSN-1, SSN-2 and classical mutagenesis methods, with one big difference—classical mutagenesis leads to thousands of random mutations, whereas SSN-1 and SSN-2 lead to single specific mutations in a targeted gene.
Another disadvantage of classical mutagenesis methods is this necessitates a selection for plants with the intended mutations, but also plant breeders must carry out several generations of backcrossing to get rid of unwanted mutations. These two latter steps are much simpler and faster when using SSN-1 or SSN-2.
Oligonucleotide-Directed Mutagenesis
The technique oligonucleotide-directed mutagenesis (ODM) uses oligonucleotides (small molecules) into which, in a similar manner to SSN-2, a small repair template is introduced into the plant cell, which is identical to the plant’s genetic material—except for the desired change.
After the DNA repair process, plants are selected where the modification has been copied into the DNA. The difference with SSN-2 is no genetic construct is copied into the DNA of the plant itself. The small repair molecule that is used remains briefly in the plant cell and is quickly degraded (Figure 2). This method only works in plants that can be regenerated from protoplasts.

It is important to mention that with SSN-1, SSN-2 and ODM, additional genetic variation is created within an existing species without crossing any species barrier. It is this creation of additional genetic variation that is absolutely crucial and fundamental to plant breeding.
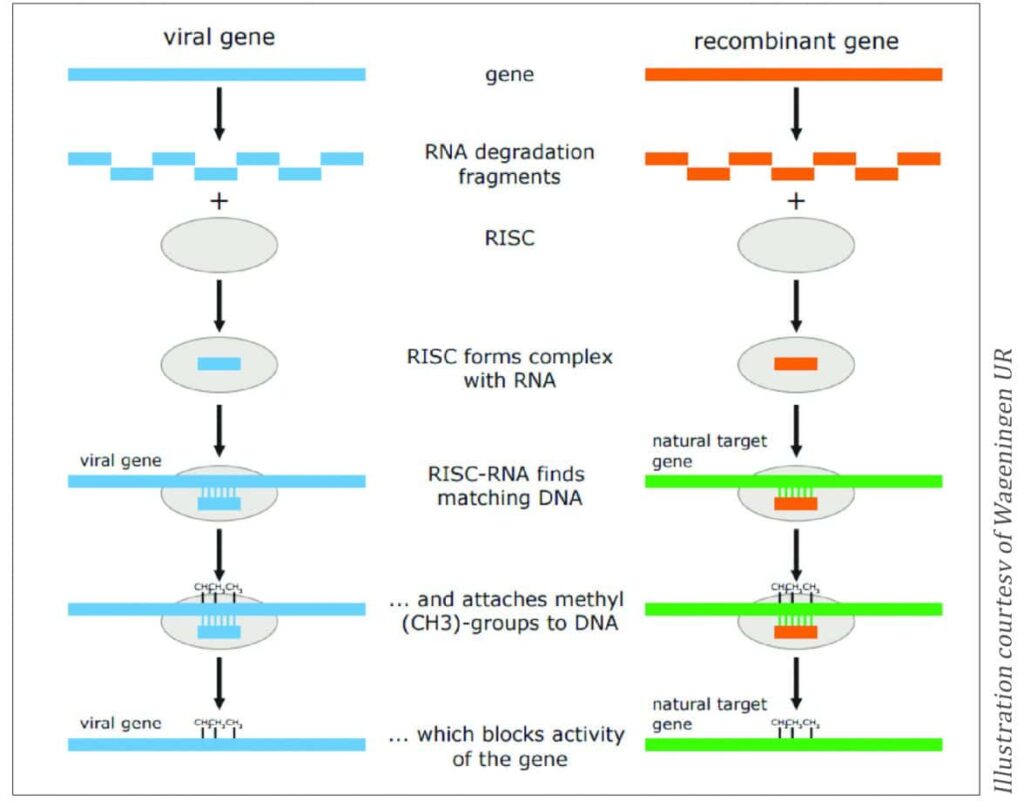
RNA-Dependent DNA methylation
RNA-dependent DNA methylation (RdDM) relies on the plant’s defence system (RNA-induced silencing complex, RISC), which is activated by small double-stranded RNA molecules (from viruses, for example). The system forms a complex with the RNA of foreign origin and methylates the matching DNA, ultimately blocking the expression of the gene (Figure 3).
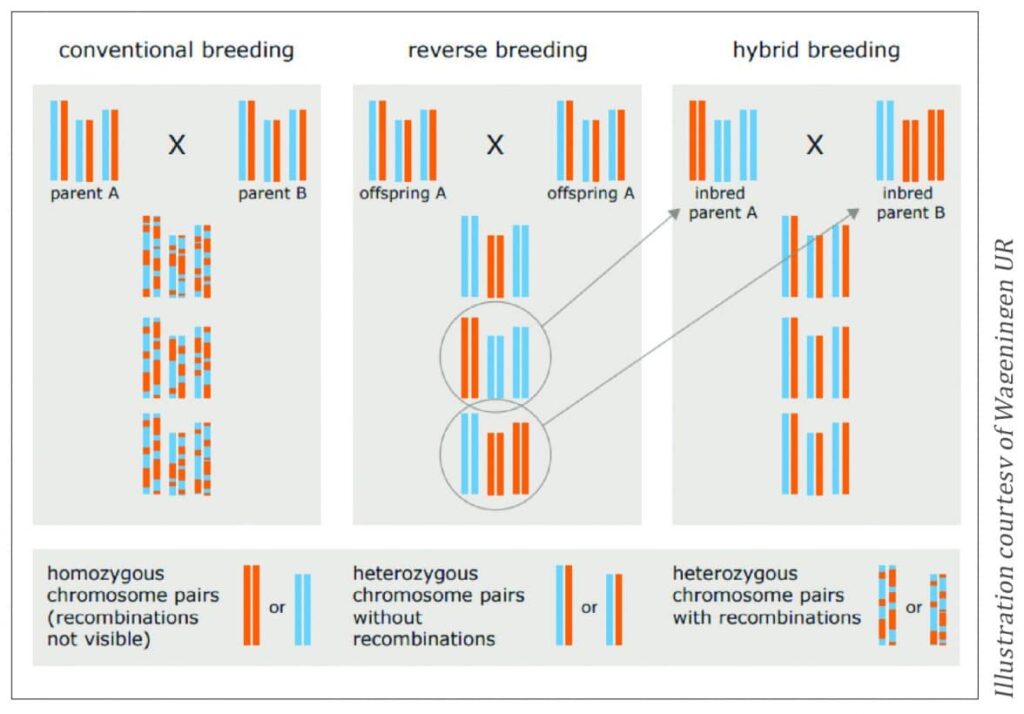
Reverse Breeding
It is not possible to exactly reproduce a heterozygous plant via seeds. Only vegetative reproduction will allow for an exact copy. However, seed companies are geared to reproduce and commercialise their elite plant varieties by means of seeds, as vegetative reproduction is often too expensive, technically cumbersome, and commercialization is often logistically impossible. Reverse breeding uses a genetic modification step to suppress the recombination of chromosomes, followed by specific tissue culture to create homozygous parent lines. These lines are then used to stably produce the heterozygous elite plants through seed (Figure 4).
GM Rootstock Grafting
With this technique, the top part of a plant, called the scion, is grafted onto a GM rootstock (Figure 5). The resulting combined plant is usually regarded as a GM plant, but the products, such as the flowers or the fruits that are harvested on the non-GM scion, do not carry the genetic modification, and are considered GM-free. This is particularly useful in cases where the rootstock conveys beneficial characteristics to the combination, such as more efficient nutrient uptake from the soil, better rooting ability in heavy soils, or resistance to soil-borne diseases (e.g. nematodes).

Induced Early Flowering
With this approach, recombinant genes are introduced into a plant that promote flowering in the first year. This is particularly helpful in trees that have a long juvenile phase in which they don’t flower. The early flowering enables much faster breeding and selection in these species. In the final breeding step, the recombinant early flowering genes are crossed out, resulting in varieties that are free of any transgenes. The plants produced in this way are indistinguishable from varieties obtained through conventional breeding, but are now achieved decades earlier.
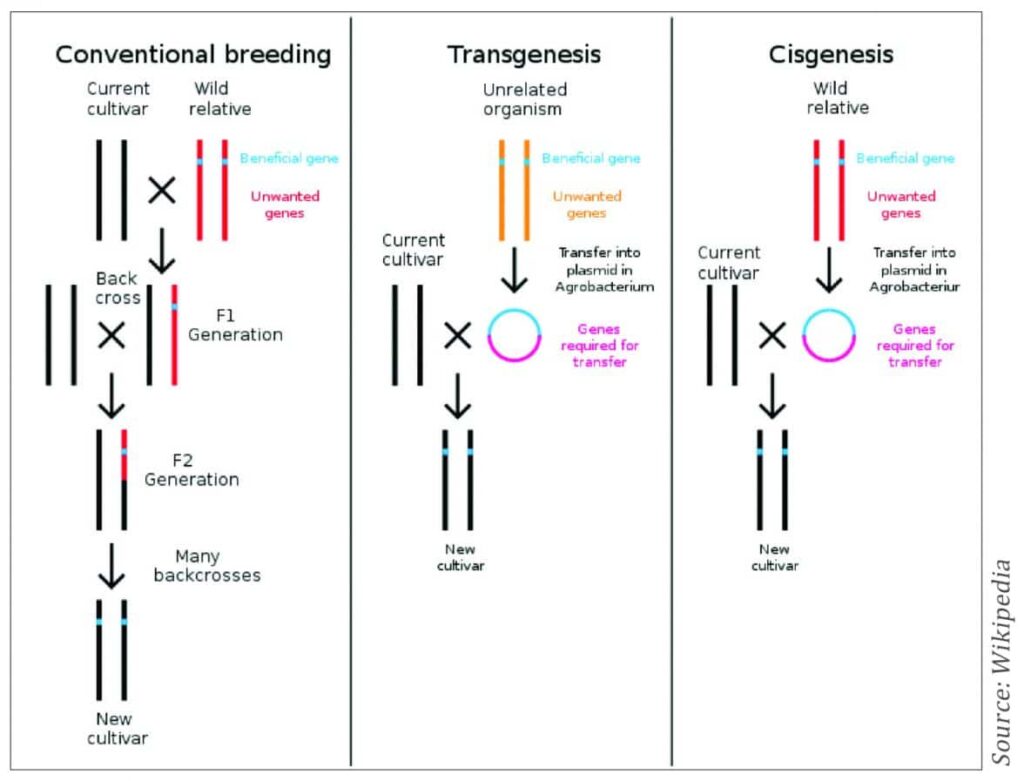
Cisgenesis
Using this method, specific traits of interest are introduced into a plant using genetic modification, but only using genes from the same species, or from a species that can be crossed with it (Figure 6). The introduced cisgene is an identical copy of a gene from the sexually compatible pool, including promoter, introns and terminator. The donor plant must be crossable with the recipient plant. This means that the same result could be achieved through conventional plant breeding, but this would take a much longer period, often up to four or five times longer. This technique is especially promising for the development of plant varieties of vegetatively propagated crops, such as potato, apple, banana, cassava and grape.
Intragenesis
Plants created with this technique contain new genes that originate from the species itself or from a crossable species. Intragenesis allows in vitro recombination of genetic elements isolated from different genes within the sexually compatible gene pool. With cisgenesis, genes are a new combination of genetic elements that cannot be obtained with traditional breeding. For example, one can replace the natural promotor by a promotor from another gene that comes from the same species.
As with every technique there are limitations. One of the limitations shared by both cisgenesis and intragenesis is traits outside the sexually compatible gene pool cannot be introduced. Additionally, the creation of intragenic crops requires new expertise and more time compared with transgenic crops. The desired genes or fragments of genes may not be readily available, but have to be isolated from the sexually compatible gene pool.
Also, the production of selection marker-free plants often requires the implementation or development of new methods, since such methods may not be readily available for the crop. This means considerable efforts have to be spent, especially on crops with low transformation efficiencies, to produce high numbers of modified plants.
The disadvantages described above for intragenesis and cisgenesis are greatly compensated by their potential to overcome some of the limitations of conventional plant breeding. Both cisgenesis and intragenesis confer a faster and more precise tool for the transfer of genetic constructs between related species than classical backcross breeding. At the same time, the linkage drag often seen in conventional backcross programs is avoided.
The intra-/cisgenic techniques can also overcome limitations of classical breeding when it comes to improving traits with limited natural allelic variation. A higher expression level of a trait can be obtained through cisgenesis by inserting an additional gene copy of the trait, or through intragenesis by introducing a hybrid gene containing an advantageous promoter and terminator isolated from the sexually compatible gene pool. Lower expression levels can be obtained through intragenesis by the introduction of different silencing constructs.
Valuable New Tools
New plant breeding techniques significantly reduce the time and effort needed to produce new plant varieties, and allow more precision. It would seem that considering the immense challenges ahead, the NBTs provide valuable new tools to EU plant breeders that are much needed in light of both the EU’s agricultural sector constraints and the global challenges concerning population increase, climate change, food security, and the sustainable use of resources.
Potential Applications of NBTs
- Potatoes with reduced amylase content and with late blight resistance.
- Apples with scab resistance and with decreased allergenicity.
- Rice with bacterial leaf blight resistance.
- Oilseed rape with herbicide tolerance.
- Wheat with powdery mildew resistance.
- Maize with drought tolerance and herbicide tolerance.
- Soybean with improved oil quality.
Advantages of NBTs
- Increased precision and efficiency of the plant breeding process.
- Arrive quicker at the desired plant characteristics.
- Faster ways to increase plants’ resistance to pests and diseases. This, in turn, leads to a reduction in the use of pesticides.
- Faster ways to increase plants’ tolerance to abiotic stresses, such as drought, leading to better use of water and other resources. With that, new plant varieties provide for a greater harvest security and higher food security.
- Overall, NBTs provide benefits to EU consumers, and have a positive impact on the environment. Food is produced in a more sustainable manner.
- More high-quality plant varieties are available for EU farmers, giving them the possibility to produce food and feed in a more efficient and sustainable way. It also provides farmers with the necessary means to generate some much-needed economic benefits.
- The techniques will become a major driver of Europe’s economy and ensure that EU plant breeders remain competitive on a
global scale.