This new department is designed to bring more scientific information to readers, as well as showcase the work being done by graduate students. This issue’s showcase focuses on stacking resistance alleles to increase resistance to soybean cyst nematode.
- Lillian Brzostowski is a Ph.D. student at the University of Illinois at Urbana-Champaign studying crop science. Her research is focused on soybean breeding and genetics. Her advisor is Dr. Brian Diers. Brzostowski received her bachelor’s and master’s in agronomy from Kansas State University.
Abbreviations:
- Chromosome (chr)
- Hexadecyltrimethylammonium
- bromide (CTAB)
- Female index (FI)
- Heterodera glycines Ichinohe (HG)
- Second stage juvenile (J2)
- Linkage group (LG)
- Polymerase chain reaction (PCR)
- Polyvinyl chloride (PVC)
- Quantitative trait loci (QTL)
- Simple sequence repeat (SSR)
Soybean cyst nematode (SCN), Heterodera glycines (HG) Ichinohe, is the pathogen that causes the greatest yield losses to soybean [Glycine max (L.) Merrill] with estimated losses in the United States of more than a billion dollars a year. The best method to manage SCN is through the use of resistant varieties. Genetic resistance to SCN is controlled by several genes known as quantitative trait loci (QTL). More than 118 soybean accessions have been identified as possible sources of resistance to SCN; however, only seven resistant sources are commonly used by breeders. The most widely used resistance source is PI88788. With the ability of some field SCN populations to overcome these resistance sources, it is important to continue to evaluate novel resistance sources. A genetic region from PI567516C on chromosome (chr) 10 (formerly linkage group (LG) O) has been identified and confirmed to confer resistance to many SCN isolates. Additionally, two resistance QTL from wild soybean (Glycine soja Siebold & Zucc.) accession PI468916 have been mapped to regions on chr 15 (LG E) and 18 (LG G). The two QTL have been confirmed and designated as cqSCN-006 and cqSCN-007, respectively. In this study, a population segregating for resistance from PI567516C, PI468916, and PI88788 was evaluated with two nematode isolates, HG type 1.2.3.5.6.7 and HG type 1.2.3.4.5.6.7. In these stacks of resistance genes, the SCN resistance alleles from each source significantly increased SCN resistance compared to the alternative alleles. Lines homozygous for the four resistance alleles had a lower SCN female index (FI) than those homozygous for the susceptible alleles. These results indicate that combining multiple sources of resistance can be an effective means to increase SCN resistance.
Introduction
Soybean cyst nematode (SCN), Heterodera glycines Ichinohe, is the greatest pathogen threat to soybean yields in the United States. From 2006-2009, the estimated average annual yield loss to SCN was 128.6 million bushels, which represents a loss of more than a billion dollars a year for producers (Koenning and Wrather, 2010). SCN was first identified in North Carolina in 1953; however, since this time, the pathogen has spread to most major soybean producing areas. In the SCN life cycle, the first stage juvenile develops within an egg and molts to become a second stage/infective stage juvenile (J2). The J2 moves a short distance through the soil to the root tips. It penetrates the root and establishes a feeding site called a syncytium and engorges. The juvenile molts three more times before becoming an adult. Females become immobile and continue to feed on the root. Their bodies swell and become yellow, lemon-shaped cysts that protrude from the root containing approximately 100 to 200 eggs. Eggs only develop if they are fertilized by a male, and at death, the cysts are brown and dislodge from the root into the soil. Adult males are vermiform and mobile, and they will stop feeding, exit the root to fertilize females, and die. The life cycle takes about 25 to 40 days with several generations occurring in a single growing season (Triantaphyllou and Hirshmann, 1962; Jardine and Todd, 2001).
While rotation to non-host crops such as corn can be a means of SCN control, it does not completely eradicate the pathogen from the soil due to SCN’s ability to overwinter in soil for several years (Jackson et al., 2005; Miller et al., 2006; Porter et al., 2001). Therefore, the best method to manage SCN is resistant cultivars. During the past several decades, SCN resistance has been a primary focus of soybean breeders (Kopisch-Obuch et al., 2005). In resistant cultivars, the nematode is able to penetrate the root and form syncytia, the nematode’s feeding site; however, the syncytia either form slowly or become necrotic soon after they are formed. This causes the nematode to starve to death (Williamson and Hussey, 1996).
Soybean genetic resistance to SCN was identified shortly after the pathogen was discovered in the United States. Caldwell et al. observed resistance from PI548402 (‘Peking’) in the form of three recessive and independent alleles: rhg1, rhg2, and rhg3 (Caldwell et. al, 1960). In a later study, Matson and Williams (1965) reported a fourth dominant resistance QTL (quantitative trait loci) from ‘Peking’, Rhg4 (Matson and Williams, 1965). Rao-Arelli et al. (1992) reported another dominant allele in PI88788, which was given the designation Rhg5 (Rao-Arelli et al., 1992).
More than 118 soybean accessions have been identified as possible sources of resistance to SCN (Arelli et al., 2000). While it appears genetic diversity for SCN resistance exists, it is narrow with only seven resistant sources commonly used by breeders. As an additional limitation, several of these sources share one or more genes. The most commercially used source of resistance is PI88788, which has broad level of resistance to HG (Heterodera glycines) types (Niblack et al., 2008) and also allows for good agronomic traits (Diers and Arelli, 1999; Shier, 2008). In an Illinois survey of SCN-resistant cultivars, more than 94 percent had resistance derived from PI88788 alone (Shier, 2008).
An SCN population is characterized using the HG type test. In this test, a profile for the population is developed based on its ability to reproduce on a standard panel of seven indicator soybean lines that confer SCN resistance. For example, an HG type 1.2 population would have elevated reproduction on indicator lines 1 (Peking) and 2 (PI88788). Prior to the HG type test, a race test was used (Niblack et al., 2002).
With the heavy reliance of breeders on just a few sources of resistance, it is important to continue to search for novel resistance sources in order for resistance to continue to be an effective means of SCN control. Young et al (1999) identified PI567516C as the only source of resistance to LY1 nematodes, a highly virulent synthetic population derived from a mass mating of HG 1.2.3 females with HG 1.2 males. Further research confirmed the resistance in the accessions (Arelli and Young, 2005) and its genetic distinction from another commonly used resistance source, ‘Hartwig’ (Chen et al., 2006). QTL were mapped using SSR (simple sequence repeat) markers in a population derived from a PI567516C x Hartwig cross. A genetic region on chr 10 (formally linkage group (LG) O), defined by the markers Satt592, Satt331, and Sat_274, was associated with LY1 resistance (Arelli et al., 2010). The chr 10 QTL conferred resistance to HG types 2.5.7, 0, 2.7, 1.3.5.6.7, and LY1 (Vuong et al. 2010). The genetic markers can be used to help select for nematode resistance.
Another way to find new resistance genes is by screening soybean relatives such as Glycine soja. Wang et al. (2001) identified a significant QTL allele for SCN resistance from G. soja PI468916 on chr 15 (LG E) and chr 18 (LG G). The QTL on chr 15 and chr 18 were later confirmed and designated as cqSCN-006 and cqSCN-007, respectively (Kabelka et al., 2005).
Further testing on the effect of these QTL indicated no major concerns of linkage drag that would reduce yield (Kabelka et al., 2006). CqSCN-006 was fine mapped to a 803.4 kb region and CqSCN-007 was mapped to a 146.5 kb region (Kim and Diers, 2013). The abundance of genetic and phenotypic information on these QTL make them good candidates to put into other genetic backgrounds to determine whether they would be appropriate for widespread use in a breeding program.
The objective of this study was to test the effect of SCN on a soybean background segregating for rhg1, the chr 15 and chr 18 QTL from PI468916, and the chr 10 resistance QTL from PI567516C.
Materials and Methods
Population Development
In 2010, LDXGL10-030-2 was backcrossed to 09SCNPOP11-9. This cross was designated as LDXGE11-288 and was fixed for rhg1 from PI88788, fixed for the chr 15 and chr 18 QTL from G. soja, and heterozygous for the PI567516C QTL.
In summer 2011, BC2F1 plants of LDXGE11-288 were crossed to LD09-15628, a BC4-derived line with the susceptible rhg1 allele fixed in a LD00-3309 background. This cross was designated the number LDX11-319 and was segregating for rhg1, the PI567516C QTL, and the two G. soja QTL. Seed from LDX11-319 was advanced in the greenhouse and field to form a F5-derived population comprised of 105 soybean lines segregating for rhg1, the PI567516C QTL, and the chr 15 and chr 18 QTL from PI468916.
Genetic Analysis
Genomic DNA was isolated from young trifoliate leaves of F4 or F5 plants via a modified CTAB method to identify the genotypic classes in the population (Keim and Shoemaker, 1998). Simple sequence repeat (SSR) markers (Cregan et al., 1999; Song et al., 2004; Song et al., 2010) linked to the QTL were used to perform polymerase chain reaction (PCR) according to the method described by Cregan and Quigley (1996). Product from PCR was separated on 6 percent (w/v) nondenaturing polyacrylamide gels by electrophloresis (Wang et al., 2003).
SCN Bioassay
SCN bioassays were performed by infesting individual F5:6 plants with HG 1.2.3.5.6.7 and 1.2.3.4.5.7 field isolates obtained from Alison Colgrove at the University of Illinois, Urbana, Illinois. The bioassays were conducted in the greenhouse in a thermo-regulated water bath system using modified methods described by Arelli et al. (2000) and Niblack et al. (2002). Briefly, PVC (polyvinyl chloride) tubes filled with a 1:1 sand-to-soil mix were placed in a plastic crock. Each tube was inoculated with approximately 2,000 nematode eggs suspended in water, and a single seedling was planted in each tube. The crock was suspended in a water bath at 27°C. The plants were watered as needed and grown under 16-hour daylength. After the test was maintained for 28 days, the tubes were soaked in water to remove the plants from the soil. Cysts were dislodged from the root on a nested 850-µm aperture over 250-µm aperture sieve using gentle water pressure and then counted using a steriomicroscope. A female index (FI) was calculated on a per plant basis using the following equation (Golden et al., 1970): FI = 100 x (Number of cysts per plant/Average number of cysts on susceptible host Lee 74).
Statistical Analysis
Data were subject to analysis using SAS v9.4 (SAS Institute Inc., Cary, NC) PROC MIXED.
Results and Discussion
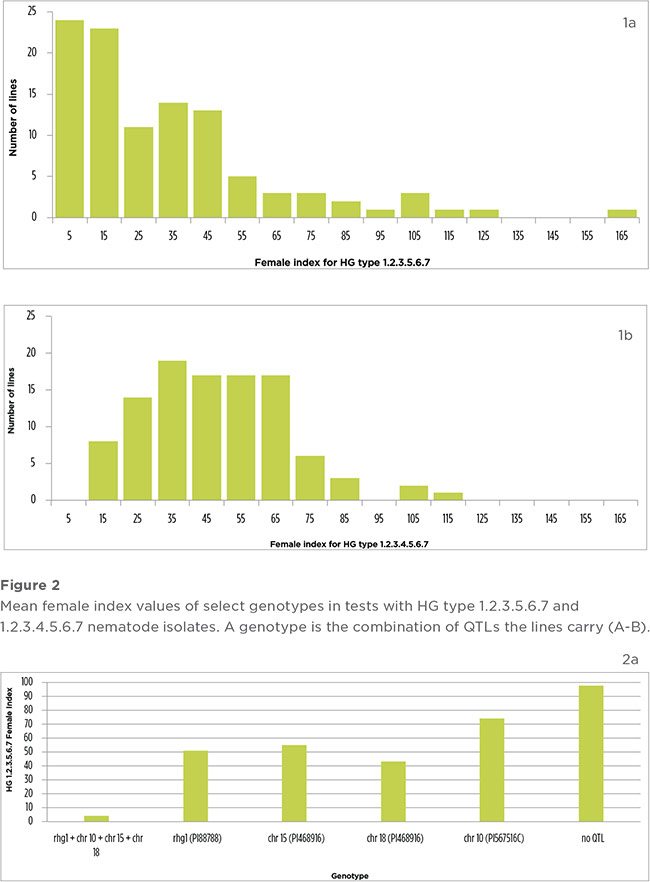
The nematode isolates used in this study were naturally occurring. HG 1.2.3.5.6.7 was collected from a field in Dekalb, Illinois, while HG 1.2.3.4.5.6.7 originated from a Missouri field population. Soybean cyst nematode reproduction on the susceptible check Lee 74 averaged 402 cysts per plant in the test with HG 1.2.3.5.6.7 and 371 cysts per plant in the test with HG 1.2.3.4.5.6.7 (Table 1). The biotypes of the SCN isolates responded as expected.
There was a continuous distribution of FI across the population for HG 1.2.3.5.6.7 and HG 1.2.3.4.5.6.7 (Figure 1a-b). Resistant lines (FI<10) were observed in the test with HG 1.2.3.5.6.7 in addition to moderately resistant (FI=10-30), moderately susceptible (FI=31-60), and susceptible lines (FI>60) (Figure 1a) (Schmitt and Shannon, 1992). In the HG 1.2.3.4.5.6.7 test, lines were moderately resistant to susceptible (Figure 1b).
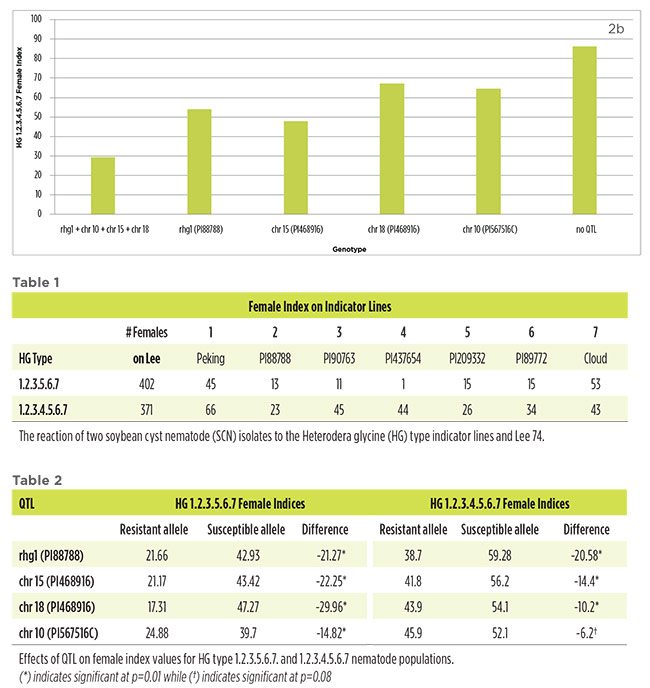
When FI data was analyzed across all of the lines (n=105), each of the four resistance QTL alleles provided partial resistance to both nematode isolates. SCN reproduction was significantly reduced with lines containing the resistance QTL alleles having a lower FI compared to lines containing the alternative QTL alleles (Table 2). The magnitude of these differences changed across QTL and SCN isolate.
On an individual line basis, lines with a single QTL had a lower FI than lines with no QTL (Figure 2a-b). The degree of these differences depended on the QTL and the SCN isolate used. Additionally, lines homozygous for all four resistance QTL alleles had reduced SCN reproduction compared to lines that only had a single QTL or lines that had no QTL. Lines homozygous for all four resistance QTL were resistant to HG 1.2.3.5.6.7 (Figure 2a). Considering that lines with only a single QTL were moderately susceptible or susceptible to the isolate, this indicates that stacking the resistance sources can be an effective means to enhance resistance to SCN. Lines homozygous for all four resistance QTL were moderately resistant to HG 1.2.3.4.5.6.7 (Figure 2b).
The soybean population used in this study was developed in order to test the effect of various combinations of rhg1 from PI88788, the chr 15 and chr 18 QTL from PI468916, and the chr 10 resistance QTL from PI567516C. The SCN isolates used were selected for their ability to overcome several of commonly used resistance sources.
Genetic resistance remains important for controlling yield losses due to SCN. PI468916 and PI567516C are alternative sources breeders can use to enhance and diversify SCN resistance in their programs. Stacking QTL from multiple resistance sources can provide resistance or partial resistance to highly virulent nematode isolates.
Future Research Direction
Select lines from the population will continue to be evaluated with additional SCN isolates, including isolates that are more commonly found in producers’ fields to continue to test if stacking the resistance sources will result in broad-level resistance. Eventually, agronomic traits of the lines will also have to be measured to ensure the QTL can be stacked without a negative effect on yield.
Acknowledgements
This research was supported with funding from the United Soybean Board.
References
Arelli, P. R., Sleper, D. A., Yue, P., & Wilcox, J. A. (2000). Soybean Reaction to Races 1 and 2 of Heterodera glycines. Crop science, 40(3), 824-826.
Arelli, P. R., Concibido, V. C., & Young, L. D. (2010). QTLs associated with resistance in soybean PI567516C to synthetic nematode population infecting cv. Hartwig. Journal of Crop Science and Biotechnology, 13(3), 163-167